New study on the genesis and spread of a novel HPAI H5N1 clade 2.3.4.4b reassortant genotype (EA-2023-DG)
Published on May 15, 2025, by Simon Dellicour
In Europe, highly pathogenic avian influenza (HPAI) virus continues to circulate in avian wildlife and undergo frequent reassortment, sporadic introductions in domestic birds, and spillover to mammals. An H5N1 clade 2.3.4.4b reassortant, EA-2023-DG, affecting wild and domestic birds was detected in western Europe in November 2023. Six of its RNA segments came from the EA-2021-AB genotype, but the PB2 and PA segments originated from low pathogenicity avian influenza viruses. In this new study, we analysed the genesis and spread of this reassortant genotype in western Europe. Discrete phylogeographic analyses of concatenated genomes and single PA and PB2 segments suggested a reassortment event in the summer of 2023 near the southwestern Baltic Sea. Subsequent continuous phylogeographic analysis of all concatenated EA-2023-DG genomes highlighted circulation in northwestern Europe until June 2024 and long-distance dispersal toward France, Norway, England, Slovakia, Switzerland, and Austria. Those results illustrate the value of phylodynamic approaches to investigate emergence of novel avian influenza virus variants, trace their subsequent dispersal history, and provide vital clues for informing outbreak prevention and intervention policies. Read the whole study here.
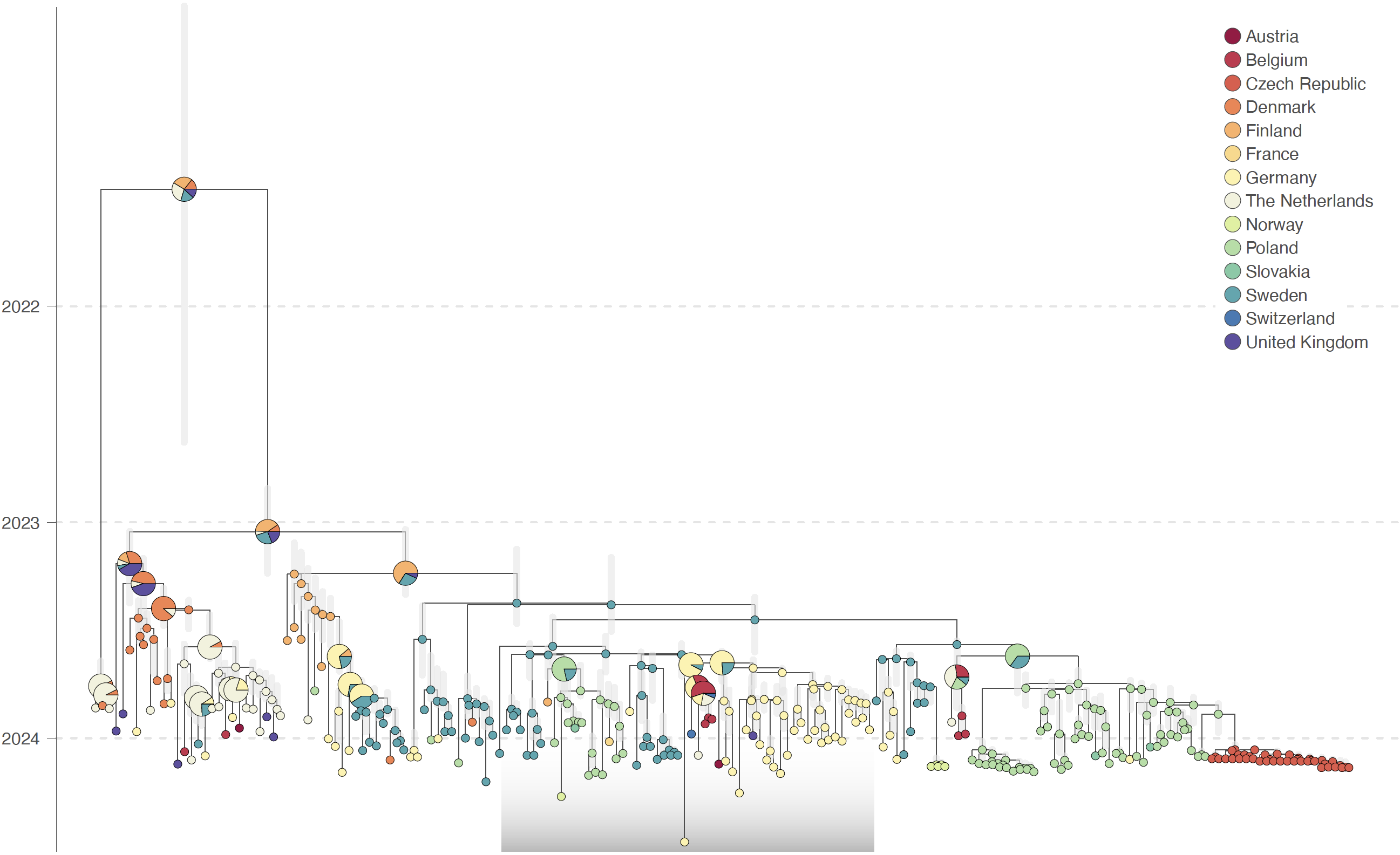 Figure: discrete phylogeographic analysis of the emergence of AIV H5N1 genotype EA-2023-DG, here based on sequences of the PA segment. We here report the time-scaled maximum clade credibility (MCC) tree obtained from the discrete phylogeographic inference based on the PA alignment, with vertical transparent gray line segments reflecting the 95% highest posterior density (HPD) associated with each internal node age estimate, as well as internal and tip nodes colored according to the inferred sampling and location, respectively. For the internal nodes, when there is not a single location inferred with a posterior probability >0.95, we use a pie chart to display the posterior probabilities associated with inferred locations with at least a posterior probability >0.05. The gray transparent boxplot highlights the position of the EA-2023-DG clade. See Figures 1, Appendix 1 Figure 1 for the discrete phylogeographic reconstruction based on the PB1-HA-NP-NA-MP-NS concatenated and PB2 alignments, respectively.
Figure: discrete phylogeographic analysis of the emergence of AIV H5N1 genotype EA-2023-DG, here based on sequences of the PA segment. We here report the time-scaled maximum clade credibility (MCC) tree obtained from the discrete phylogeographic inference based on the PA alignment, with vertical transparent gray line segments reflecting the 95% highest posterior density (HPD) associated with each internal node age estimate, as well as internal and tip nodes colored according to the inferred sampling and location, respectively. For the internal nodes, when there is not a single location inferred with a posterior probability >0.95, we use a pie chart to display the posterior probabilities associated with inferred locations with at least a posterior probability >0.05. The gray transparent boxplot highlights the position of the EA-2023-DG clade. See Figures 1, Appendix 1 Figure 1 for the discrete phylogeographic reconstruction based on the PB1-HA-NP-NA-MP-NS concatenated and PB2 alignments, respectively.
Reference: Van Borm S, Ahrens AK, Bachofen C, Banyard A, Bøe CA, Briand FX, Dirbakova Z, Engelsma M, Fusaro A, Germeraad E, Gjerset B, Grasland B, Harders F, Hostyn P, Kauppinen A, Lambrecht B, Mollett B, Monne I, Nagy A, Pohlmann A, Polzer D, Reid S, Revilla-Fernandez S, Steensels M, Stätter M, Swieton E, Tammiranta N, Wyler M, Zecchin B, Zohari S, Dellicour S (2025). Genesis and spread of novel highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus genotype EA-2023-DG reassortant, western Europe. Emerging Infectious Diseases, in press