New study on the dispersal and human-fish host switching history of Streptococcus agalactiae ST283
Published on October 04, 2023, by Dan Schar and Simon Dellicour
Fish consumption-associated outbreaks of Streptococcus agalactiae (group B Streptococcus; GBS) sequence type (ST) 283 in Asia have drawn attention to GBS ST283 as an emerging foodborne pathogen capable of generating disease in the general population. To inform public health interventions, researchers gathered 328 whole genome sequences collected from humans and fish between 1998 and 2021 across eleven countries spanning four continents, applying Bayesian modeling to reconstruct the evolutionary history of ST283, host transitions and geographic dispersal. Findings suggest expansion within Asia following emergence in the early 80s, with subsequent intercontinental dispersal over the last 20 years to the Americas. The study identified frequent bidirectional human-fish host switching, with more human-to-fish than fish-to-human transitions, suggesting transmission cycles could be interrupted though improved sanitation and wastewater management. Increased clinical awareness, enhanced surveillance and improved typing and sequencing of invasive GBS isolates are needed to inform a broader understanding of transmission risk — particularly in underserved populations reliant on small-scale farmed freshwater fish for nutrition — and through community and foodborne transmission pathways. Read the whole study here.
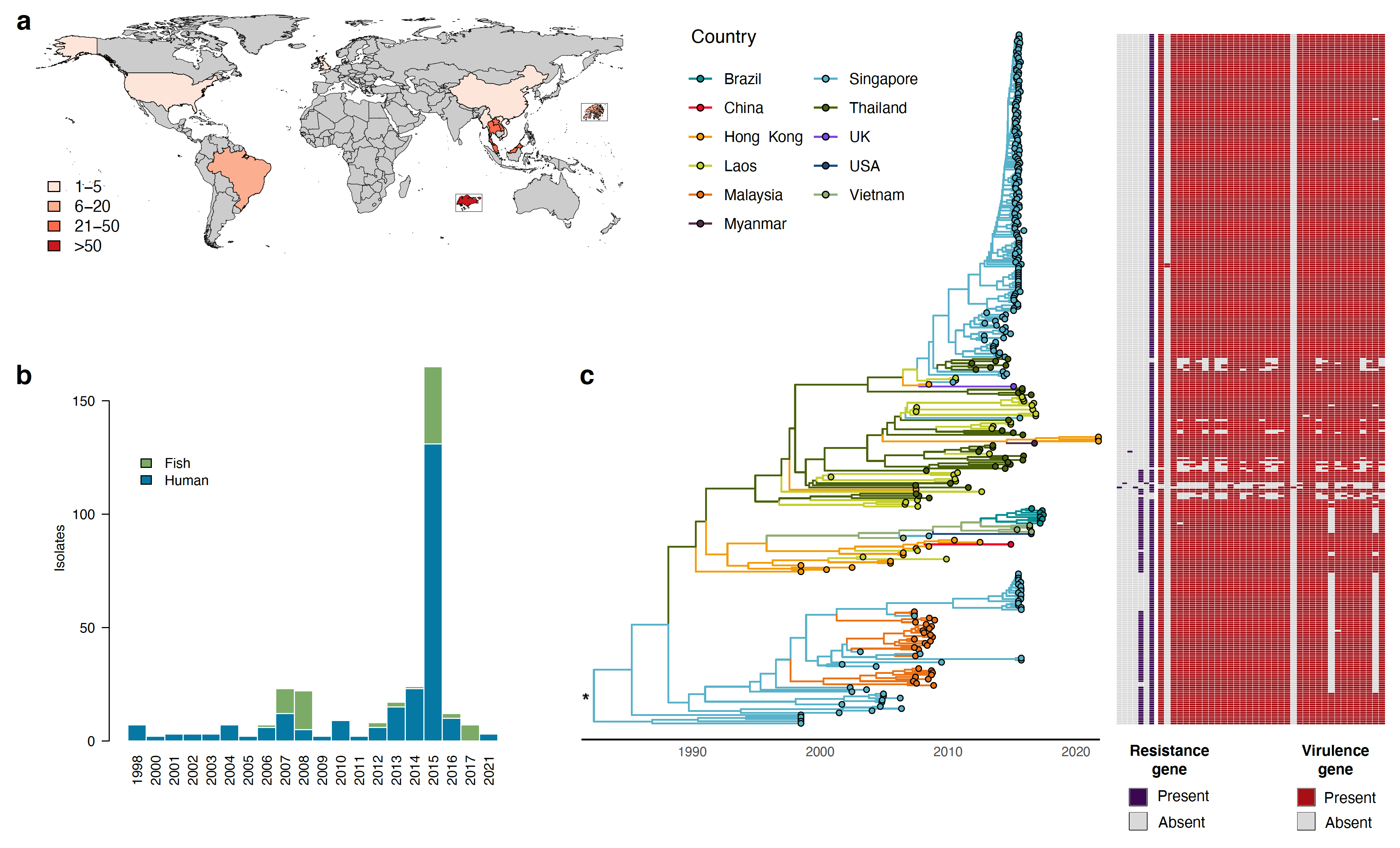
Figure: spatio-temporal distribution of GBS ST283 isolates and inferred time-scaled phylogenetic tree. (a) Map of sampling location for ST283 isolates in the study (n = 328). Insets are Singapore (bottom) and Hong Kong (right). (b) Distribution of isolate sampling dates and host origin. (c) Annotated maximum clade credibility (MCC) tree resulting from a discrete phylogeographic analysis. Tip nodes and branches are colored according to the country of origin and the country inferred at ancestral nodes. Asterisk (*) indicates that, after having taken the heterogeneity of the sampling effort among sampled locations into account, no meaningful support was identified for ancestral root node location (see the Results section for further detail). A table of antimicrobial resistance (blue) and virulence factor (red) gene presence or absence is displayed for each isolate in the study.
Reference: Schar D, Zhang Z, Pires J, Vrancken B, Suchard MA, Lemey P, Ip M, Gilbert M, Van Boeckel T, Dellicour S (2023). Dispersal history and bidirectional human-fish host switching of invasive, hypervirulent Streptococcus agalactiae sequence type 283. PLoS Global Public Health 3: e0002454