New publication on methodological developments in landscape genetics
Published on March 01, 2019, by Simon Dellicour
Our article titled “Landscape genetic analyses of Cervus elaphus and Sus scrofa: comparative study and analytical developments” has recently been published in Heredity. In this study, we describe and use a landscape genetic workflow to compare spatial patterns of genetic variability and the impact of environmental factors on genetic differentiation.
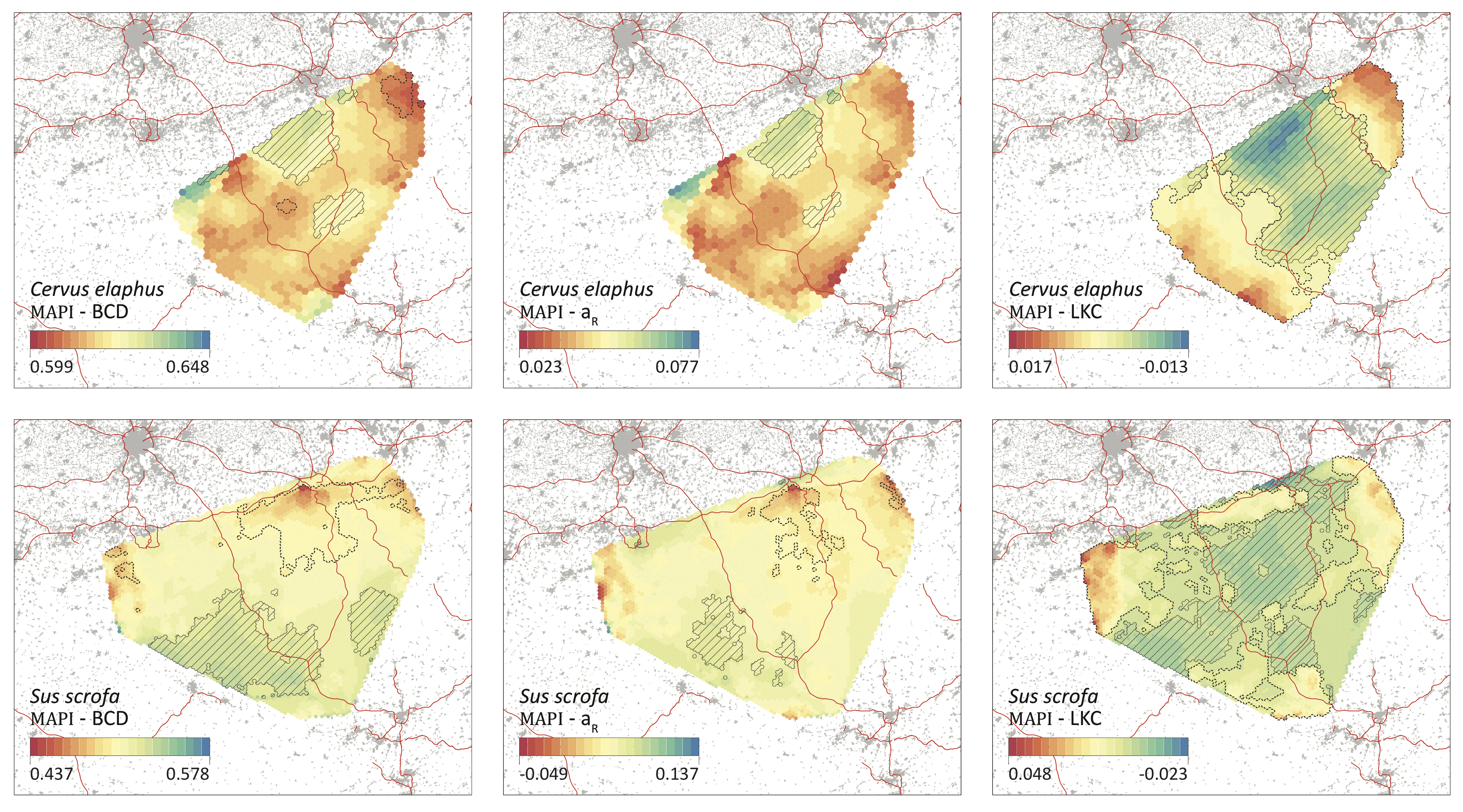
Red deer and wild boar are two major game species whose populations are managed and live in areas impacted by human activities. Measuring and understanding the impact of landscape features on individual movements and spatial patterns of genetic variability in these species is thus of importance for managers. A large number of individuals sampled across Wallonia (Belgium) for both species have been genotyped using microsatellite markers (respectively >1700 and >1200 genotyped individuals) and some individuals have also been followed using a capture-mark-recapture (CMR) protocol. The combined data set represents an unprecedented opportunity to study and compare the environmental factors impacting the interconnectivity of these large mammals. The present study describes and uses a landscape genetic workflow to compare spatial patterns of genetic variability and the impact of environmental factors on genetic differentiation. For the latter analyses, we investigate the correlation between genetic and environmental distances (pairwise approach) and also between local genetic dissimilarity and environmental conditions (point approach). Preliminary analyses of CMR data confirm that motorways act as significant barriers to dispersal. However, analyses performed with the pairwise approach do not highlight any evidence of an impact of motorways on genetic differentiation, which is presumably due to their recent establishment. Complementary analyses performed with the point approach reveal that low altitude tends to be associated with higher genetic dissimilarity. From a methodological point of view, our workflow illustrates the complementary application of both pairwise and point approaches as well as univariate and multivariate analyses. Read the whole study here.
Reference: Dellicour S, Prunier JG, Piry S, Eloy MC, Bertouille S, Licoppe A, Frantz AC, Flamand MC (2019). Landscape genetic analyses of Cervus elaphus and Sus scrofa: comparative study and analytical developments. Heredity 123: 228-241