Uncovering the endemic circulation of rabies in Cambodia - our new study published in Molecular Ecology
Published on August 17, 2023, by Simon Dellicour
Our new study on the rabies virus circulation in Cambodia has been published in Molecular Ecology. In epidemiology, endemicity characterises sustained pathogen circulation in a geographical area, which involves a circulation that is not being maintained by external introductions. Because it could potentially shape the design of public health interventions, there is an interest in fully uncovering the endemic pattern of a disease. Here, we use a phylogeographic approach to investigate the endemic signature of rabies virus (RABV) circulation in Cambodia. Cambodia is located in one of the most affected regions by rabies in the world, but RABV circulation between and within Southeast Asian countries remains understudied. Our analyses are based on a new comprehensive data set of 199 RABV genomes collected between 2014 and 2017 as well as previously published Southeast Asian RABV sequences. We show that most Cambodian sequences belong to a distinct clade that has been circulating almost exclusively in Cambodia. Our results thus point toward rabies circulation in Cambodia that does not rely on external introductions. We further characterise within-Cambodia RABV circulation by estimating lineage dispersal metrics that appear to be similar to other settings, and by performing landscape phylogeographic analyses to investigate environmental factors impacting the dispersal dynamic of viral lineages. The latter analyses do not lead to the identification of environmental variables that would be associated with the heterogeneity of viral lineage dispersal velocities, which calls for a better understanding of local dog ecology and further investigations of the potential drivers of RABV spread in the region. Overall, our study illustrates how phylogeographic investigations can be performed to assess and characterise viral endemicity in a context of relatively limited data. Read the whole study here.
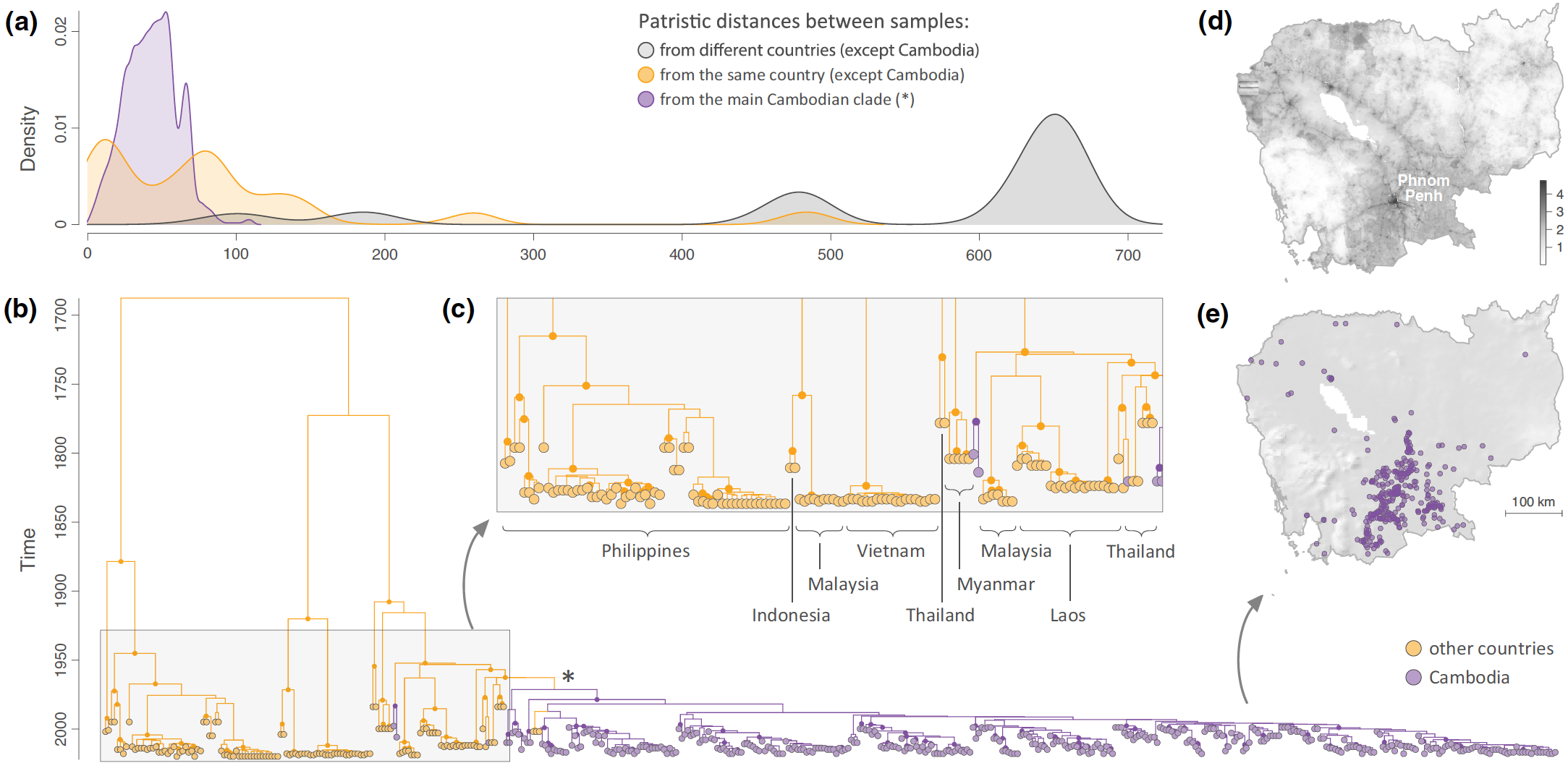
Investigating the endemic signature of RABV circulation in Cambodia. (a) Comparison between patristic distances (in years) computed on the maximum clade credibility (MCC) at the inter-countries level (excluding Cambodia; in grey), at the intra-country level (excluding Cambodia; in orange), and within the main Cambodian clade (in purple). (b) MCC tree obtained from the preliminary discrete phylogeographic inference based on the N gene sequences and considering only two different discrete locations: ‘Cambodia’ and ‘other countries’. Tip nodes and phylogeny branches are coloured according to their sampling location and inferred ancestral location, respectively. On the MCC tree, we only highlight the internal nodes associated with a posterior probability >0.95. The asterisk refers to the main Cambodian clade inferred by the discrete phylogeographic analysis, whose introduction in Cambodia is estimated to be around 1971 (95% HPD = [1963-1979]). (c) Zoom on one particular part of the MCC tree corresponding to the terminal branches outside the main Cambodian clade. The country of origin of sampled sequences are displayed below the tip nodes. (d) Map of log10-transformed human population density in Cambodia. (e) Sampling map of all N gene sequences used for the discrete phylogeographic analysis (n = 354).
Reference: Layan M, Dacheux L, Lemey P, Brunker K, Ma L, Troupin C, Dussart P, Chevalier V, Wood J, Ly S, Duong V, Bourhy H, Dellicour S (2023). Uncovering the endemic circulation of rabies in Cambodia. Molecular Ecology 32: 5140-5155