Diseases risk mapping
Studying livestock diseases
Our initial research has dealt with avian influenza (“bird flu”) H5N1 and H7N9 in Asia. In 2004, the highly pathogenic avian influenza (HPAI) H5N1 virus started spreading throughout Asia, causing the death of millions of poultry and transmitting occasionaly to humans, with some fatalities. In the following years, the virus spread across Eurasia down to Africa. Today, it persists in a limited set of countries (Egypt, Bangladesh, China, Vietnam, Indonesia). Our work consisted in identifying the main risk factors associated with the presence or absence of the diseases at multiple scale, in particular the role of host species, and type of farming. Recently, a different avian influenza H7N9 virus emerged in China, low pathogenic in poultry this time, but causing human fatalities too.
Our research has aimed to understand the agro-ecological drivers of avian influenza emergence. Why are these viruses emerging? Where are they more likely to emerge? Why are they showing a higher apparent seasonality? What is the respective role of wild bird connectivity and trade in pattern of virus migrations? These are the numerous questions we are trying to tackle through geospatial analysis and modelling.
Our work also entails producting suitability maps at different spatial scale that can be used to be target suveillance and control in high-risk areas. In the long run, we aim to better understand the role of changes in production and trade systems in the emergence and spread of those viruses, so that we can project how changes in those systems resulting from intensification of the production may translate into disease risk. In addition, we aim to better complement geospatial approaches with phylogeographic analysis and inference to better understand how different factors may shape the evolution and spread of these viruses.
Previous work has involved studies on bovine tuberculosis in Belgium and in the UK, and on the vectors of Bluetongue disease in Belgium and Italy. We have worked on applying spatial modelling to different disease such as Bluetongue in Europe, Porcine Reproductive and Respiratory Syndrome (PRRS), Foot and Mouth Disease (FMD) and Nipah virus infections in Thailand.
Impact of global change
More recently, the SpELL has been leading studies dedicated to the analyses of the impact of climate and land-use changes on the distribution of pathogens of OneHealth importance such as the Lassa virus in Africa and the West Nile virus (WNV) in Europe. For instance, we have investigated the extent to which WNV spatial expansion in Europe can be attributed to climate change while accounting for other direct human influences such as land use and human population changes. To this end, we have trained ecological niche models to predict the risk of local WNV circulation leading to human cases to then unravel the isolated effect of climate change by comparing factual simulations to a counterfactual based on the same environmental changes but a counterfactual climate where long-term trends have been removed. Our findings demonstrate a notable increase in the area ecologically suitable for WNV circulation during the period 1901-2019, whereas this area remains largely unchanged in a no-climate-change counterfactual. In this study, we also show that the drastic increase in the human population at risk of exposure is partly due to historical changes in population density, but that climate change has also been a critical driver behind the heightened risk of WNV circulation in Europe.
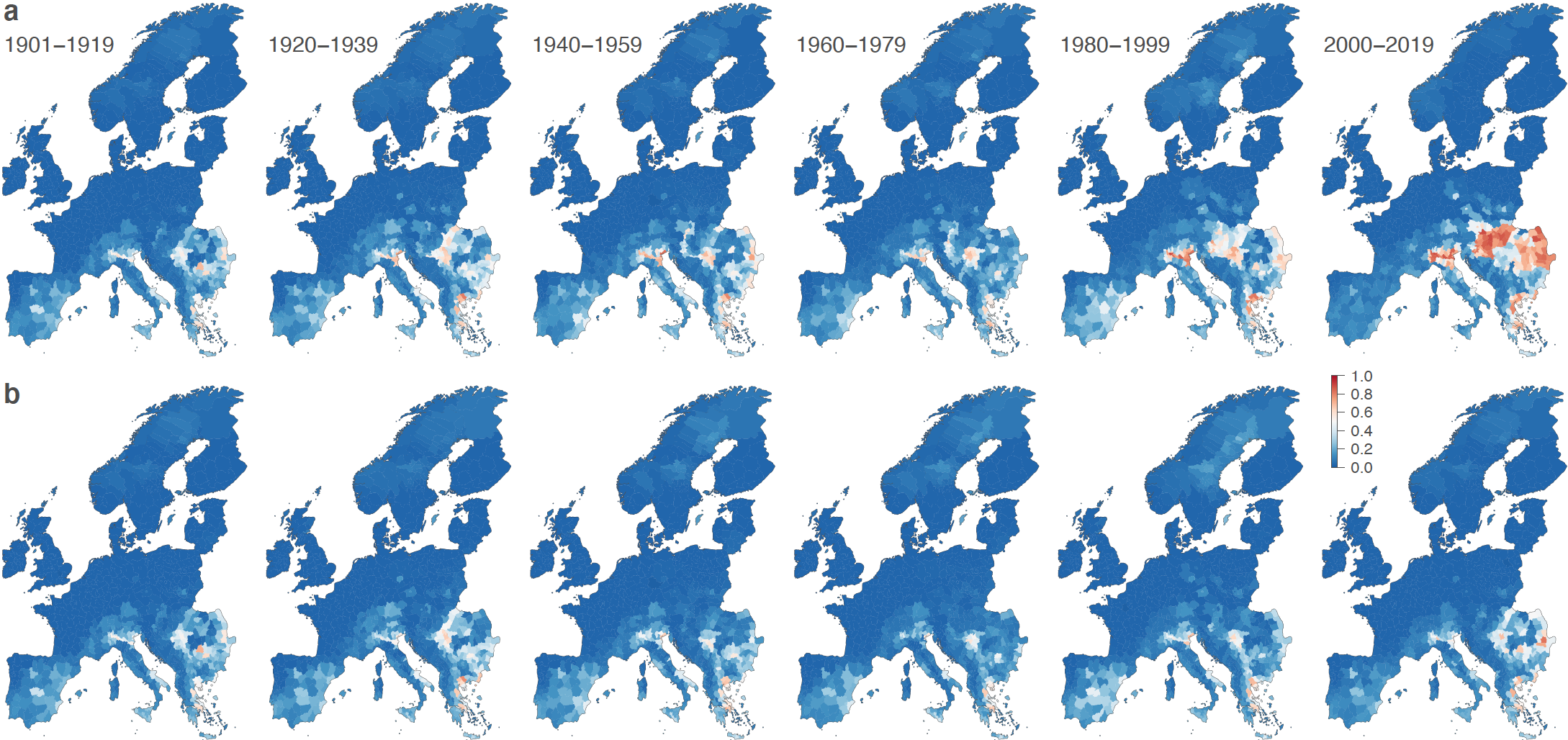
Figure: estimated changes in the risk of local West Nile virus circulation since the beginning of last century across the European continent. The successive maps display the estimated evolution of ecological suitability based on actual environmental data (a) and a counterfactual scenario representing a world without climate change (b).
Regarding the emergence of WNV in Europe, we have also integrated indicator-based and event-based surveillance data to improve risk mapping of the risk of human exposure to that virus. The ecological niche models based on both types of surveillance data highlighted new areas potentially at risk of WNV infection in humans, particularly in Spain, Italy, France and Greece, which underscored the potential for a more proactive and comprehensive strategy in managing the threat of WNV in Europe by combining indicator- and event-based and environmental data for effective surveillance and public health response.
Mapping livestock
Our lab is also leading and conducting research projects to better map the distribution of livestock production at a global scale. For many years, several research groups and international organisations have developed high-resolution global maps of livestock based on reported statistics derived from census and survey data. Due to the dynamic nature of these populations (intensification of livestock production) this is an extremely challenging task that involves the collection of contemporary statistics, the development of complex algorithms to disaggregate these statistics at higher spatial resolutions and make predictions where they are absent.

The main objectives of our ongoing work are to evaluate quantitatively and qualitatively the different approaches that are used to predict and represent livestock distribution in high-resolution global databases. Our initial efforts focused on the establishment of global livestock distribution data representing the best data available to date (Gilbert et al. 2018).
We now work on the development of time series for the past (1990 - present) and for the future (present - 2050) based on time series of predictor variables and on adapted modelling procedures that can take advantages of training data in space and time. In parrallel, we also worked on the development of different models that are better suited to predict monogastric species (poultry and pigs) that can be raised in very high number within a single locations. We investigated the use of point-pattern simulation modelling to predict farm locations (Chaiban et al. 2019) and ocne calibrated, the approach will be applied to create realistic synthetic farm distribution models in relevant countries.
Further developments involve predicting how intensification of the livestock production sector may impact the geoghraphical distribution of animals and farms in ruminant and monogastric livestock production systems. We also work toward the development of animal movement models (Nicolas et al. 2018) that can be used to predict movement networks. Our work is carried out in collaboration with the FAO Livestock and Environment group, and is presented in FAO Livestock Systems web page. The data themselves are distributed through the Global Livestock Data Harvard dataverse.
Collaborations
Our main institutional collaborators are or have been T. Robinson (ILRI, Nairobi, Kenya), G. Dauphin, W. Kalpravidth, S. Newman, V. Martin, J. Slingenbergh, G. Cinardi (FAO, Rome, Italy), W. Thanapongtharm (Department of Livestock Development, DLD, Bangkok, Thailand), H. Yu (Chinese Center for Disease Control, CDC, Beijing, China), A. Conte & Carla Ippoliti (IZS Teramo), and our main academic collaborators are or have been Xiangming Xiao (Univ. Oklahoma, USA), Julien Cappelle (CIRAD, Montpellier, France), D. Pfeiffer (Royal Veterinary College, London, UK), N. Golding, S. Hay (SEEG, Univ. Oxford, Oxford, UK), W. Wint (ERGO ltd, Oxford, UK), S. Vanwambeke (UCLouvain, Louvain-la-Neuve, Belgium), Kristian Andersen (Scripps Research, US), Raphaëlle Klitting (Aix-Marseille University, France), Wim Thiery (Vrij Universiteit Brussel, Belgium), Nuno Faria (Imperial College, London, UK), and William de Souza (University of Kentucky, US).
Selected publications
Contribution of climate change to the spatial expansion of West Nile virus in Europe
D. Erazo, L. Grant, G. Ghisbain, G. Marini, F. J. Colón-González, W. Wint, A. Rizzoli, W. Van Bortel, C. B. F. Vogels, N. D. Grubaugh, M. Mengel, K. Frieler, W. Thiery, and S. Dellicour.
"Nature Communications",
Vol. 15,
Issue 1,
Pages 1196,
2024.
Integrating indicator-based and event-based surveillance data for risk mapping of West Nile virus, Europe, 2006 to 2021
K. Serres, D. Erazo, G. Despréaux, M. F. Vincenti-González, W. V. Bortel, E. Arsevska, and S. Dellicour.
"Eurosurveillance",
Vol. 29,
Issue 44,
Pages 2400084,
2024.
Predicting the evolution of the Lassa virus endemic area and population at risk over the next decades
R. Klitting, L. E. Kafetzopoulou, W. Thiery, G. Dudas, S. Gryseels, A. Kotamarthi, B. Vrancken, K. Gangavarapu, M. Momoh, J. D. Sandi, A. Goba, F. Alhasan, D. S. Grant, S. Okogbenin, E. Ogbaini-Emovo, R. F. Garry, A. R. Smither, M. Zeller, M. G. Pauthner, M. McGraw, L. D. Hughes, S. Duraffour, S. Günther, M. A. Suchard, P. Lemey, K. G. Andersen, and S. Dellicour.
"Nature Communications",
Vol. 13,
Issue 1,
Pages 5596,
2022.
Global mapping of highly pathogenic avian influenza H5N1 and H5Nx clade 2.3.4.4 viruses with spatial cross-validation
M. S. Dhingra, J. Artois, T. P. Robinson, C. Linard, C. Chaiban, I. Xenarios, R. Engler, R. Liechti, D. Kuznetsov, X. Xiao, S. V. Dobschuetz, F. Claes, S. H. Newman, G. Dauphin, and M. Gilbert.
"eLife",
Vol. 5,
Pages e19571,
2016.
Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia
M. Gilbert, N. Golding, H. Zhou, G. R. W. Wint, T. P. Robinson, A. J. Tatem, S. Lai, S. Zhou, H. Jiang, D. Guo, Z. Huang, J. P. Messina, X. Xiao, C. Linard, T. P. Van Boeckel, V. Martin, S. Bhatt, P. W. Gething, J. J. Farrar, S. I. Hay, and H. Yu.
"Nature Communications",
Vol. 5,
Pages 4116,
2014.