Dispersal dynamics of SARS-CoV-2 lineages during the first epidemic wave in New York City
Published on May 26, 2021, by Simon Dellicour
Our new study on the dispersal dynamics of SARS-CoV-2 lineages during the first epidemic wave in New York City has been published in PLoS Pathogens. In this study, we performed phylogeographic investigations to gain insights into the circulation of viral lineages during the first months of the New York City outbreak, during which the city rapidly became the epicenter of the pandemic in the United States. Our analyses describe the dispersal dynamics of viral lineages at the state and city levels, illustrating that peripheral samples likely correspond to distinct dispersal events originating from the main metropolitan city areas. In line with the high prevalence recorded in this area, our results highlight the relatively important role of the borough of Queens as a transmission hub associated with higher local circulation and dispersal of viral lineages toward the surrounding boroughs. Read the whole study here.
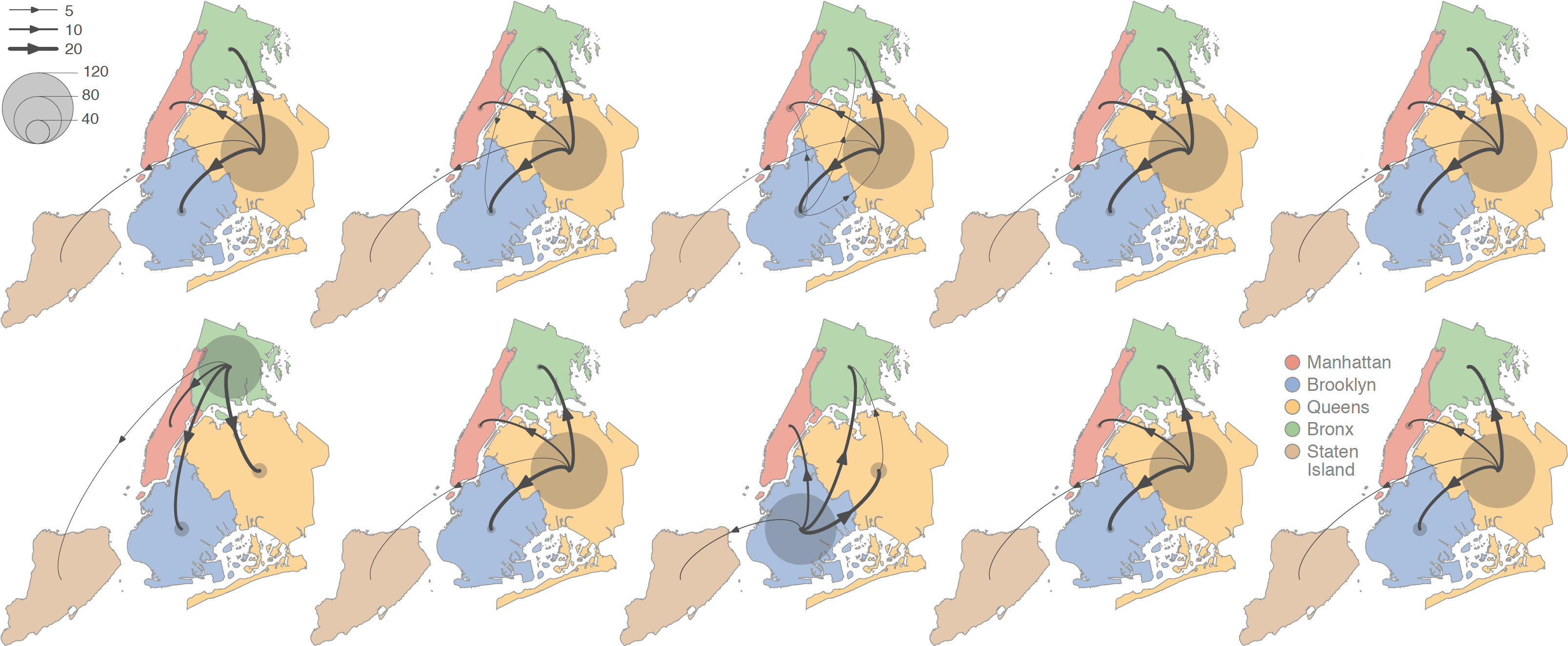
Figure 3: Schematic overview of the discrete phylogeographic analyses of the main SARS-CoV-2 clade circulating in New York City (NYC) during the first epidemic wave. These maps schematize the outcome of each replicated discrete phylogeographic inference based on a random subset of genomic sequences (see the Methods section for further detail). We here report the number of lineage dispersal events inferred among (arrows) and within (transparent grey circles) NYC boroughs, both measures being averaged over 1,000 posterior trees sampled from each posterior distribution.
Reference: Dellicour S, Hong SL, Vrancken B, Chaillon A, Gill MS, Maurano MT, Ramaswami S, Zappile P, Marier C, Harkins GW, Baele G, Duerr R, Heguy A (2021). Dispersal dynamics of SARS-CoV-2 lineages during the first epidemic wave in New York City. PLoS Pathogens 17: e1009571