Our new study about epidemiological hypothesis testing using phylogeographic and phylodynamic approaches
Published on September 21, 2020, by Simon Dellicour
Our new study on landscape phylogeography has been accepted for publication in Nature Communications. In this study, we illustrate how phylogeographic and phylodynamic and analyses can be leveraged for hypothesis testing in molecular epidemiology using West Nile virus in North America as an example.
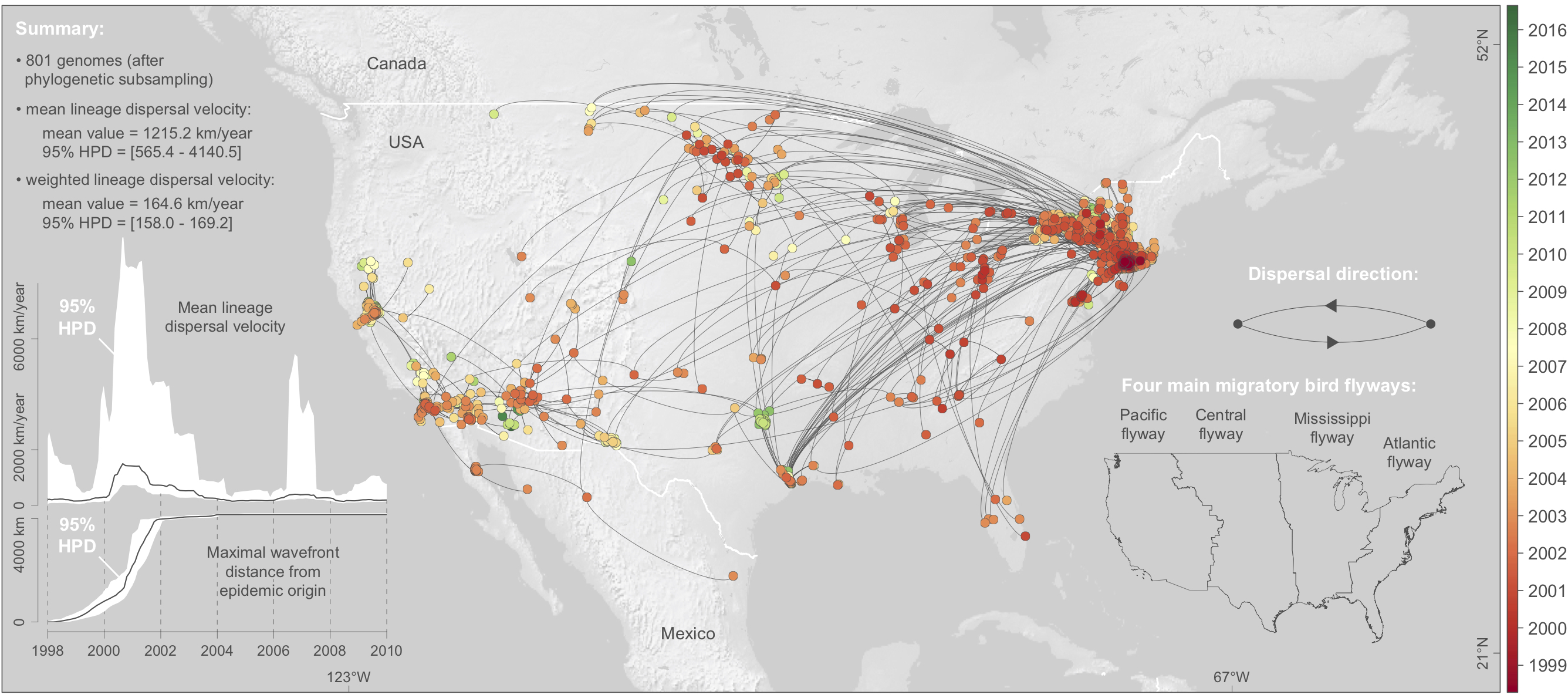
Computational analyses of pathogen genomes are increasingly used to unravel the dispersal history and transmission dynamics of epidemics. In our new study, we show how to go beyond historical reconstructions and use spatially-explicit phylogeographic and phylodynamic approaches to formally test epidemiological hypotheses. We illustrate our approach by focusing on the West Nile virus (WNV) spread in North America that has been responsible for substantial impacts on public, veterinary, and wildlife health. WNV isolates have been sampled at various times and locations across North America since its introduction to New York twenty years ago. We exploit this genetic data repository to demonstrate that factors hypothesised to affect viral dispersal and demography can be formally tested. Specifically, we detail and apply an analytical workflow consisting of state-of-the art methods that we further improve to test the impact of environmental factors on the dispersal locations, velocity, and frequency of viral lineages, as well as on the genetic diversity of the viral population through time. We find that WNV lineages tend to disperse faster in areas with higher temperatures and we identify temporal variation in temperature as a main predictor of viral genetic diversity through time. Using a simulation procedure, we find no evidence that viral lineages preferentially circulate within the same migratory bird flyway, suggesting a substantial role for non-migratory birds or mosquito dispersal along the longitudinal gradient. Finally, we also separately apply our testing approaches on the three WNV genotypes that circulated in North America in order to understand and compare their dispersal ability. Our study demonstrates that the development and application of statistical approaches, coupled with comprehensive pathogen genomic data, can address epidemiological questions that might otherwise be difficult or impractically expensive to answer. Read the whole study here.
Reference: Dellicour S, Lequime S, Vrancken B, Gill MS, Bastide P, Gangavarapu K, Matteson N, Tan Y, du Plessis L, Fisher AA, Nelson MI, Gilbert M, Suchard MS, Andersen KG, Grubaugh ND, Pybus OG, Lemey P (2020). Epidemiological hypothesis testing using a phylogeographic and phylodynamic framework. Nature Communications 11: 5620